I. Introduction
Antibiotics are usually used to enhance the production, improve feed efficiency, and treat diseases in food-producing animals to meet the challenge of providing sufficient amounts of food for the growing population (Beyene, 2016). However, antibiotics residue is one of the most important global issues concerning food contamination. It is because some bacteria are recognized as a serious public health threat due to their resistance to several antibiotics, often critically important for human health, thereby limiting the choice of effective antibiotic agents available for treatment. The existence of antibiotics residue in meat, milk, egg, and offal may result from inappropriate or extra-label usage of drugs or may not maintain withdrawal period of drugs or poor livestock production practices (Tajick and Shohreh, 2006). The actualizations of WTO regulations claim that the producer and veterinarians who work in food animal medicine should tactfully avoid drug residues in food animals and proclaim this message to the farmers for the safety of general public health. Moreover, the veterinarians employed in pharmaceutical and regulatory sectors have big responsibilities for assessing the drugs and chemicals residues that enter the human food chain through the edible products. Indeed, as most animal derived foods are being consumed after processing or cooking (Tian et al., 2016), and different cooking procedures, temperature, storage time, and the fermentation process has the potential effects to reduce veterinary drug residues (Heshmati, 2015). Therefore, the amount of antibiotics residues we consume is very low, and the problem is the emergence of antibiotics-resistant bacteria that occur because we use a lot of antibiotics, not the antibiotics we consume. This is why low dosages of antibiotics for a short duration to unhealthy dairy cattle and consumption of pasteurized milk are recommended to protect human health. For this reason, the objectives of this paper are to review the information of antibiotic in livestock and their effects on the human health.
II. Use of Antibiotic in Livestock
In an attempt to meet the protein needs of a growing human population, animal food farms became large and sought new production processes to maximize the growth rates of their animals. These increased growth and production rates helped to make food protein more available and affordable to this growing population as well as maximized profit to the producer. Increased production rates also increased the need for more livestock. An increase in livestock then led to unsanitary conditions and disease in the animals, prompting producers to administer constant low doses of antibiotics to their animals as a prophylactic to prevent disease. However, the use of antibiotics in food production also led to the spread and development of antimicrobial resistance (AMR) (Lekshmi et al., 2017). The extent to which antibiotic use in animals contributes to the antimicrobial resistance in humans is still under debate. There is already high resistance against many antibiotics used in livestock (Catry et al., 2003). However, a wealth of epidemiological information already indicates that food of animal origin is the source of a majority of food borne bacterial infections caused by Campylobacter, Yersinia, and Escherichia coli, non-typhoid Salmonella and some other pathogens (Schroeder et al., 2002).
In the case of resistance due to antibiotic use in food animals, antimicrobial resistance to bacteria occurs due to the similarity in the structure of the components in the antibiotics used in humans and animals. For example, avoparcin is used extensively in livestock animals and is structurally similar to vancomycin, which is used in hospitals as a treatment for severe infections. The result of over usage and similarity to one another led to the emergence and spread of vancomycin resistant enterococci (VRE) in hospitals. This is a specific drug resistance but the cross-recognition and cross-resistance that occur with resistant bacteria allow them to be resistant to most, if not all, antimicrobial therapies within the same class, some of which are important for human therapy. Bacteria rapidly undergo what is called horizontal gene transfer (HGT), which is how a new bacterium acquires resistance. During HGT bacteria pass between each other mobile genetic elements containing antimicrobial resistant genes (ARG) in addition to other functional genes. This along with the rapid process of HGT allows for expeditious microorganism growth and passage of genetic components and advantageous mutations (Franklin et al., 2016). Development of resistance is the result of changes, including inactivation or modification of the antibiotic by enzymes within the cell, absence of or bypassing of an enzymatic or metabolic step targeted by the antibiotic as well as impaired uptake or efflux of the antibiotic, modification of the antibiotic target site or overproduction of molecule (Schwarz and Chaslus-Dancla, 2001).
III. Animal to Human Spread of Antibiotic Resistance
Farm and slaughterhouse workers, veterinarians, and those in close contact with farm workers are directly at risk of being colonized or infected with resistant bacteria through close contact with colonized or infected animals (Fig. 1). Although this limited transmission does not initially appear to pose a population-level health threat, occupational workers and their families provide a conduit for the entry of resistance genes into the community and hospital environments, where further spread into pathogens is possible (Molbak et al., 1999). The majority of studies examining the transmission of antibiotic-resistant bacteria from animals to farm workers document the prevalence of resistance among farmers and their contacts or among farmers before and after the introduction of antibiotics at their workplace. Direct spread of bacteria from animals to people was first reported by Levy et al. (1976), who found the same tetracycline-resistant E. coli strains in the gut flora of chicken caretakers as in the chickens receiving tetracycline laced feed. The observation extended to the farm family as well and showed an increased frequency of tetracycline resistant and multidrug-resistant E. coli after several months of use of AGP-laden feed. Studies such as this (which examined a variety of antibiotic classes and assorted pathogens) have consistently shown a higher prevalence of resistant gut bacteria among farm workers than in the general public or among workers on farms not using antibiotics (Aubry-Damon et al., 2004; Katsunuma et al., 2007). While gentamicin is ot approved for growth promotion in the United States, it remains the most commonly used antibiotic in broiler production, being employed for prevention of early poultry mortality (Luangtongkum et al., 2006). A revelatory 2007 study found that the risk for carrying gentamicin-resistant E. coli was 32 times higher in poultry workers than in other members of the community: half of all poultry workers were colonized with gentamicin-resistant E. coli, while just 3% of nonpoultry workers were colonized. Moreover, the occupationally exposed population was at significantly greater risk for carriage of multidrug-resistant bacteria (Price et al., 2007). New gene-based methods of analysis provide even stronger evidence for the animal origin of bacteria that colonize or infect humans. Homologous relationships between bacterial resistance genes in humans and farm animals have been identified most commonly for food-borne pathogens such as E. coli and Salmonella (see below) but have also been recorded for various species of Enterococcus and for methicillin-resistant Staphylococcus aureus (MRSA). Zhang and colleagues found E. coli strains resistant to apramycin (an antibiotic used in agriculture but not in human medicine) in a study of Chinese farm workers. All farms in the study that used apramycin as an AGP had workers that carried apramycin resistance genes. The same resistance gene, aac(3)-IV, was present in each swine, poultry, and human isolate, with some resistance profiles also matching across species (Zhang et al., 2009). A group of French scientists found the same resistance gene [aac(3)-IV] in cow, pig, and human E. coli strains that bore resistance to apramycin and gentamicin (Chaslus-Dancla et al., 1991). In another study, similar resistance patterns and genes were detected in E. faecalis and E. faecium strains from humans, broilers, and swine in Denmark (Aarestrup et al., 2000). Lee sampled MRSA isolates from cattle, pigs, chickens, and people in Korea and found that 6 of the 15 animal isolates containing mecA (the gene responsible for methicillin resistance in S. aureus) were identical to human isolates (Lee, 2003).
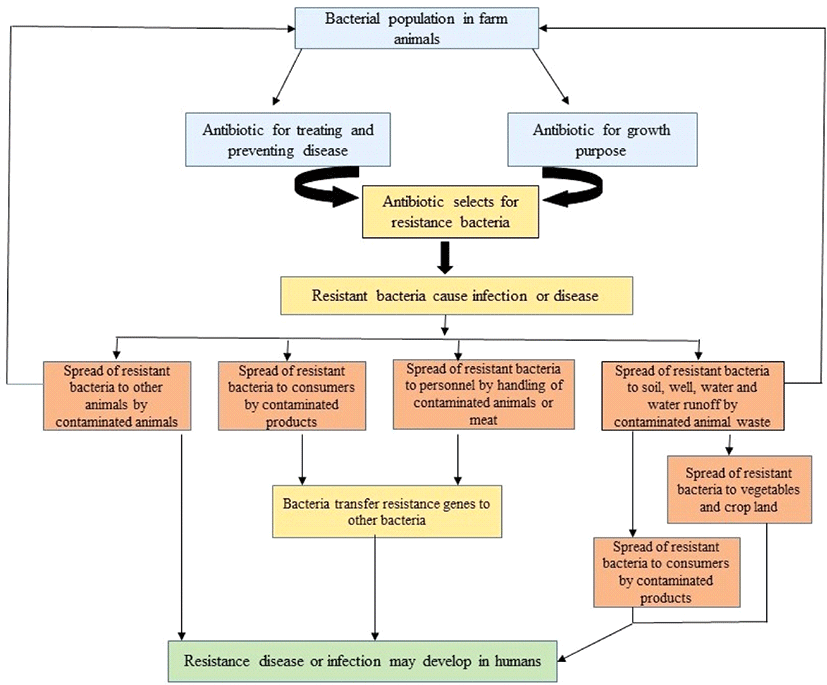
IV. Antibiotic Resistance Transmission through the Food Chain
Consumers may be exposed to resistant bacteria via contact with or consumption of animal products-a far-reaching and more complex route of transmission. There is undeniable evidence that foods from many different animal sources and in all stages of processing contain abundant quantities of resistant bacteria and their resistance genes. The rise of antibiotic-resistant bacteria among farm animals and consumer meat and fish products has been well documented (Lee, 2003; Perreten, 2005; Witte, 2000). Demonstrating whether such reservoirs of resistance pose a risk to humans has been more challenging as a consequence of the complex transmission routes between farms and consumers and the frequent transfer of resistance genes among host bacteria. Such correlations are becoming more compelling with the advent of molecular techniques which can demonstrate the same gene (or plasmid) in animal or human strains, even if the isolates are of different species. For example, Alexander et al. (2010) showed that drug-resistant E. coli was present on beef carcasses after evisceration and after 24 h in the chiller and in ground beef stored for 1 to 8 days. Others isolated ciprofloxacin-resistant Campylobacter spp. from 10% to 14% of consumer chicken products (Gupta et al., 2004; Smith et al., 1999). MRSA has been reported to be present in 12% of beef, veal, lamb, mutton, pork, turkey, fowl, and game samples purchased in the consumer market in the Netherlands (De boer, et al., 2009), as well as in cattle dairy products in Italy (Normanno et al., 2007). Likewise, extensive antibiotic resistance has been reported for bacteria, including human pathogens, from farmed fish and market shrimp (Heuer et al., 2009). Some of the antibiotic resistance genes identified in food bacteria have also been identified in humans, providing indirect evidence for transfer by food handling and/or consumption. In 2001, Sorensen et al. confirmed the risk of consuming meat products colonized with resistant bacteria, showing that glycopeptide-resistant Enterococcusfaecium of animal origin ingested via chicken or pork lasted in human stool for up to 14 days after ingestion (Sorensen et al., 2001). Donabedian et al. found overlap in the pulsed-field gel electrophoresis (PFGE) patterns of gentamicin-resistant isolates from humans and pork meat as well as in those of isolates from humans and grocery chicken (Donabedian et al., 2003). They identified that when a gene conferring antibiotic resistance was present in food animals, the same gene was present in retail food products from the same species. Most resistant enterococci possessed the same resistance gene, aac (6)-Ieaph (2)-Ia (Dumonceaux et al., 2006).
V. Emergence of Resistance in Human Infections
There is likewise powerful evidence that human consumption of food carrying antibiotic-resistant bacteria has resulted, either directly or indirectly, in acquisition of antibiotic-resistant infections. In 1985, scientists in Arizona traced an outbreak of multidrug-resistant Salmonellaenterica serovar Typhimurium, which included the death of a 72-year-old woman, to consumption of raw milk. Isolates from most patients were identical to the milk isolates, and plasmid analysis showed that all harbored the same resistance plasmid (Tacket et al., 1985). A 1998 S. typhimurium outbreak in Denmark was caused by strains with nalidixic acid resistance and reduced fluoroquinolone susceptibility. PFGE revealed that a unique resistance pattern was common to Salmonella strains from all patients, two sampled pork isolates, the swine herds of origin, and the slaughterhouse (Molbak et al., 1999). Samples from gentamicin-resistant urinary tract infections (UTIs) and fecal E. coli isolates from humans and food animal sources in China showed that 84.1% of human samples and 75.5% of animal samples contained the aaaC2 gene for gentamicin resistance (Ho et al., 2010). Johnson et al. (2007) used PFGE and random amplified polymorphic DNA (RAPD) profiles of fluoroquinolone-resistant E. coli strains in human blood and fecal samples and in slaughtered chickens to determine that the two were virtually identical to resistant isolates from geographically linked chickens. Drug-susceptible human E. coli strains, however, were genetically distinct from poultry bacteria, suggesting that the ciprofloxacin-resistant E. coli strains in humans were imported from poultry rather than originating from susceptible human E. coli (Johnson et al., 2007). Other reports demonstrate a broader linkage of resistance genes through the farm-to-fork food chain. A resistance-specifying bla CMY gene was found in all resistant isolates of Salmonellaenterica serotype Newport originating from humans, swine, cattle, and poultry. The host plasmid, which conferred resistance to nine or more antimicrobials, was capable of transmission via conjugation to E. coli as well (Zhao et al., 2003). An observed homology between CMY-2 genes in cephalosporin-resistant E. coli and Salmonella suggested that plasmids conferring resistance had moved between the two bacterial species. The authors found higher rates of CMY-2 in strains from animals than in those from humans, supporting an animal origin for the human pathogen (Winokur et al., 2001). A 2000 study found matching PFGE profiles among vancomycin-resistant Enterococcusfaecium isolates from hospitalized humans, chickens, and pigs in Denmark. Molecular epidemiology studies have also linked tetracycline resistance genes from Aeromonas pathogens in a hospital effluent to Aeromonas strains from a fish farm (Rhodes et al., 2000). These results support the clonal spread of resistant isolates among different populations (Hammerum et al., 2000). Chronologic studies of the emergence of resistance across the food chain also strongly imply that reservoirs of resistance among animals may lead to increased resistance in consumers of animal food products. Bertrand et al. (2006) chronicled the appearance of the extended-spectrum beta-lactamase (ESBL) gene CTX-M-2 in Salmonellaenterica in Belgium. This resistance element was identified first in poultry flocks and then in poultry meat and, finally, human isolates (Bertrand et al., 2006). A recent Canadian study also noted a strong correlation between ceftiofur-resistant bacteria (the pathogen Salmonellaenterica serovar Heidelberg and the commensal E. coli) from retail chicken and human infections across Canada. The temporary withdrawal of ceftiofur injection from eggs and chicks dramatically reduced resistance in the chicken strains and the human Salmonella isolates, but the trend reversed when the antibiotic use was subsequently resumed (Dutil et al., 2010). In three countries (United States, Spain, and the Netherlands), a close temporal relationship has been documented between the introduction of fluoroquinolone (sarafloxacin and enrofloxacin) therapy in poultry and the emergence of fluoroquinolone-resistant Campylobacter in human infections. An 8- to 16-fold increase in resistance frequency was observed—from 0 to 3% prior to introduction to 10% in the United States and the Netherlands and to 50% in Spain—within 1 to 3 years of the licensure (Endtz et al., 1991; Smith et al., 1999). In the Netherlands, this frequency closely paralleled an increase in resistant isolates from retail poultry products (Endtz et al., 1991), while the U.S. study used molecular subtyping to demonstrate an association between the clinical human isolates and those from retail chicken products (Smith et al., 1999). It is now theorized, from molecular and epidemiological tracking, that the resistance determinants found in Salmonella outbreaks (strain DT104) in humans and animals in Europe and the United States likely originated in aquaculture farms of the Far East. The transmissible genetic element contains the florfenicol gene (floR) and the tetracycline class G gene, both of which were traced to Vibrio fish pathogens (Vibriodamsel and Vibrioanguillarum, respectively). Both drugs are used extensively in aquaculture (Cabello, 2006).
